| Data source |
GEO: GSE158849
|
| Description |
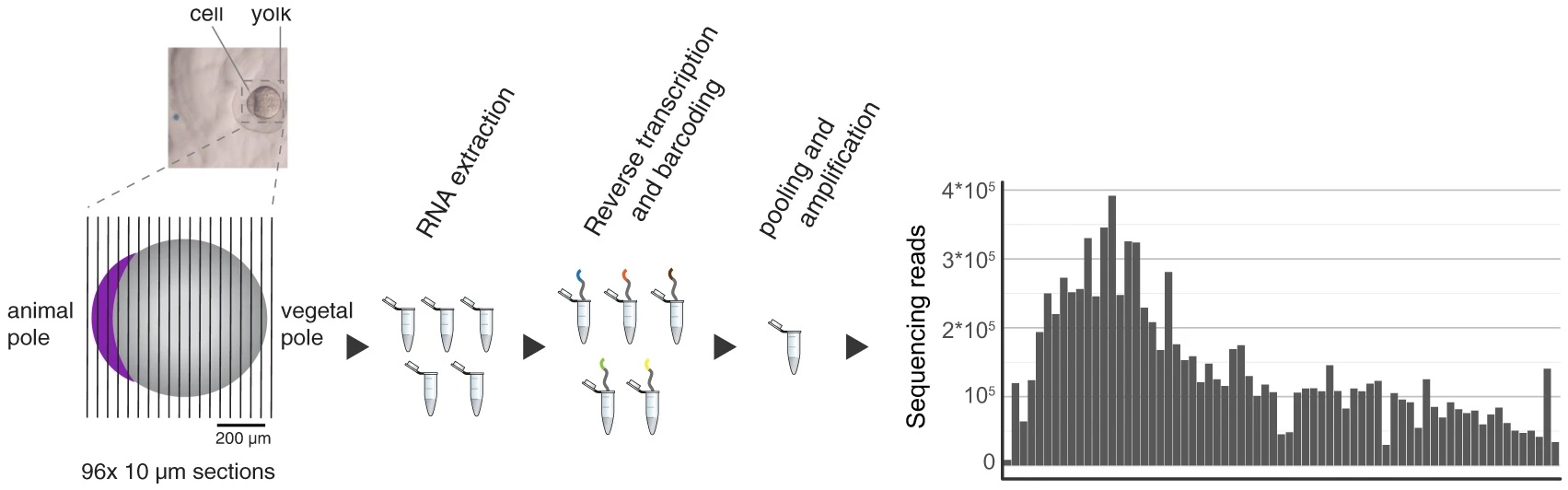
We performed spatially resolved transcriptomics with sub-single-cell resolution in zebrafish embryos at the one-cell stage, which allowed us to identify a class of mRNAs that is specifically localized at an extraembryonic position in the yolk sac, the vegetal pole. The 3’ UTRs of these localized genes are enriched in specific sequence motifs. Comparison to two frog species revealed relatively low conservation of localized genes, but high conservation of sequence motifs. In vivo RNA labeling followed by scRNA-seq revealed that a large number of the localized transcripts are specifically transported to the primordial germ cells. |
| Key word |
genome-wide expression; spatial transcriptomics; xenopus oocytes; global analysis; seq reveals; localization; translation; cells; translocation; tomography |
| Publication |
Holler K, Neuschulz A, Drewe-Boß P, Mintcheva J et al. Spatio-temporal mRNA tracking in the early zebrafish embryo. Nat Commun 2021 Jun 7;12(1):3358. PMID: 34099733 |
| Abstract |
Early stages of embryogenesis depend on subcellular localization and transport of maternal mRNA. However, systematic analysis of these processes is hindered by a lack of spatio-temporal information in single-cell RNA sequencing. Here, we combine spatially-resolved transcriptomics and single-cell RNA labeling to perform a spatio-temporal analysis of the transcriptome during early zebrafish development. We measure spatial localization of mRNA molecules within the one-cell stage embryo, which allows us to identify a class of mRNAs that are specifically localized at an extraembryonic position, the vegetal pole. Furthermore, we establish a method for high-throughput single-cell RNA labeling in early zebrafish embryos, which enables us to follow the fate of individual maternal transcripts until gastrulation. This approach reveals that many localized transcripts are specifically transported to the primordial germ cells. Finally, we acquire spatial transcriptomes of two xenopus species and compare evolutionary conservation of localized genes as well as enriched sequence motifs. Early stages of embryogenesis are known to depend on subcellular localization and transport of maternal mRNA, but systematic analyses have been hindered by a lack of methods for tracking of RNA. Here the authors combine spatially-resolved transcriptomics and single-cell RNA labeling to perform a spatio-temporal analysis of the transcriptome during early zebrafish development, revealing insights into this process. |