| Data source |
GEO: GSE159709
|
| Description |
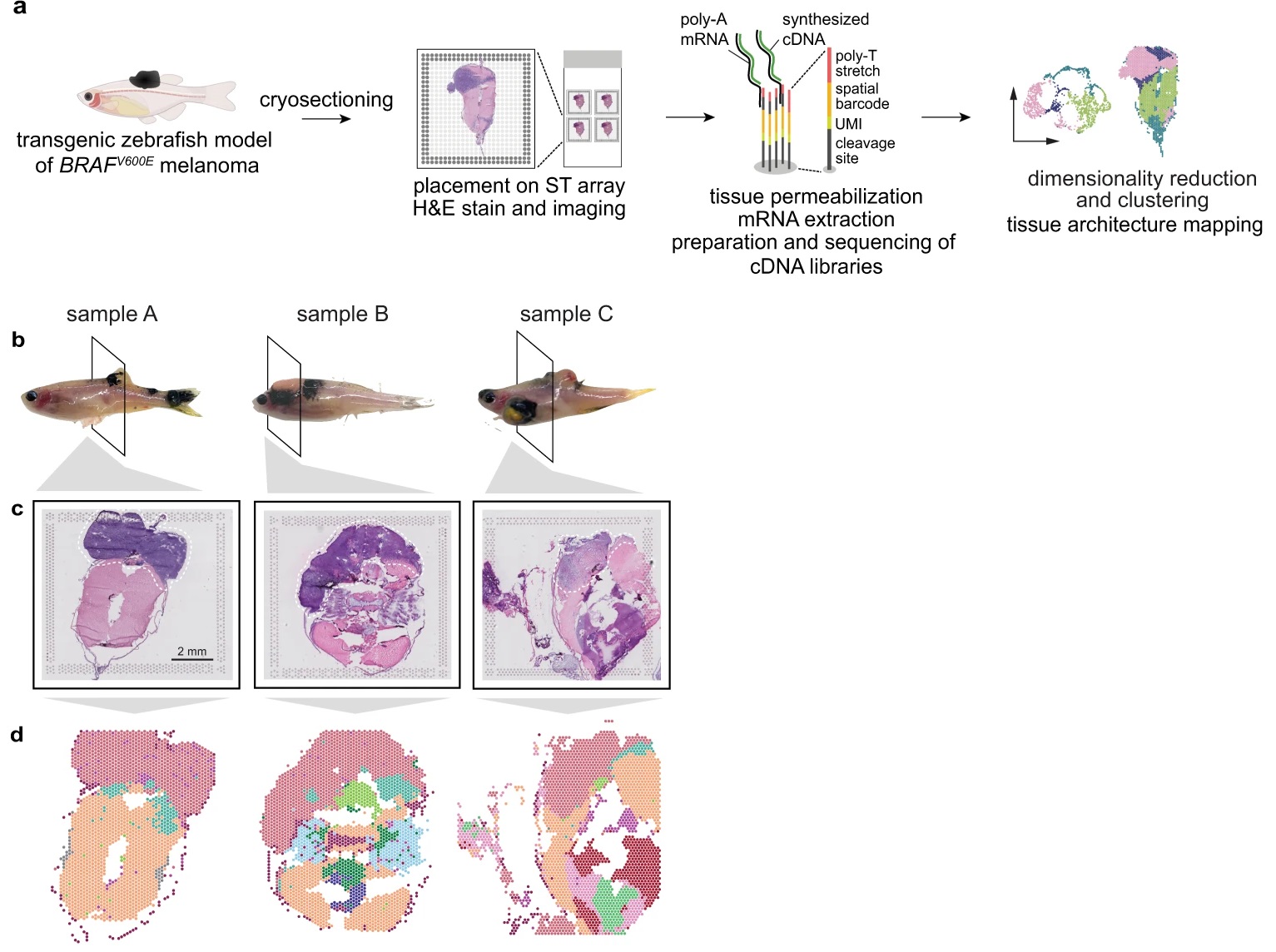
Cancer cells interact with a wide variety of other cell types, but our understanding of microenvironmental heterogeneity and how it influences tumor phenotypes is limited. While single-cell RNA-seq (scRNA-seq) has helped define these TME cell types, it provides limited information on the mechanisms that define how individual tumor cells interact with TME. Here, we integrate spatial transcriptomics with scRNA-seq to define the architecture and nature of nascent tumor and surrounding microenvironment cells as they come into contact through the process of invasion. Using a well-defined transgenic zebrafish model of BRAFV600E-driven melanoma, we identify a transcriptionally unique “interface†cluster localized at the boundary between tumor cells and surrounding tissues. Using an unbiased, data-driven approach, we identify spatially-patterned gene modules specific to the interface and show that the interface is a distinct transcriptional entity that histologically resembles the microenvironment but transcriptionally resembles the tumor. By complementing ST with scRNA-seq, we demonstrate that the interface is composed of specialized tumor and microenvironment cells. Both cell types in the interface upregulate a common set of cilia genes, and we find enrichment of cilia proteins only where the tumor meets the TME. Cilia gene expression is regulated by ETS-family transcription factors, which normally act to suppress their expression outside of this region. This unique ETS-driven interface transcriptional state is conserved across ten different human patient samples, suggesting this is a conserved feature of human melanoma. Taken together, our results demonstrate the power of spatial and single-cell transcriptomics techniques in uncovering novel biological mechanisms that drive tumor invasion into new tissues. |
| Key word |
genome-wide expression; nucleus rna-seq; gene-expression; primary cilium; single-cell; metastatic melanoma; adult zebrafish; ets family; heterogeneity; progression |
| Publication |
Hunter MV, Moncada R, Weiss JM, Yanai I et al. Spatially resolved transcriptomics reveals the architecture of the tumor-microenvironment interface. Nat Commun 2021 Nov 1;12(1):6278. PMID: 34725363 |
| Abstract |
During tumor progression, cancer cells come into contact with various non-tumor cell types, but it is unclear how tumors adapt to these new environments. Here, we integrate spatially resolved transcriptomics, single-cell RNA-seq, and single-nucleus RNA-seq to characterize tumor-microenvironment interactions at the tumor boundary. Using a zebrafish model of melanoma, we identify a distinct "interface" cell state where the tumor contacts neighboring tissues. This interface is composed of specialized tumor and microenvironment cells that upregulate a common set of cilia genes, and cilia proteins are enriched only where the tumor contacts the microenvironment. Cilia gene expression is regulated by ETS-family transcription factors, which normally act to suppress cilia genes outside of the interface. A cilia-enriched interface is conserved in human patient samples, suggesting it is a conserved feature of human melanoma. Our results demonstrate the power of spatially resolved transcriptomics in uncovering mechanisms that allow tumors to adapt to new environments. |