| Data source |
GEO: GSE104057
|
| Description |
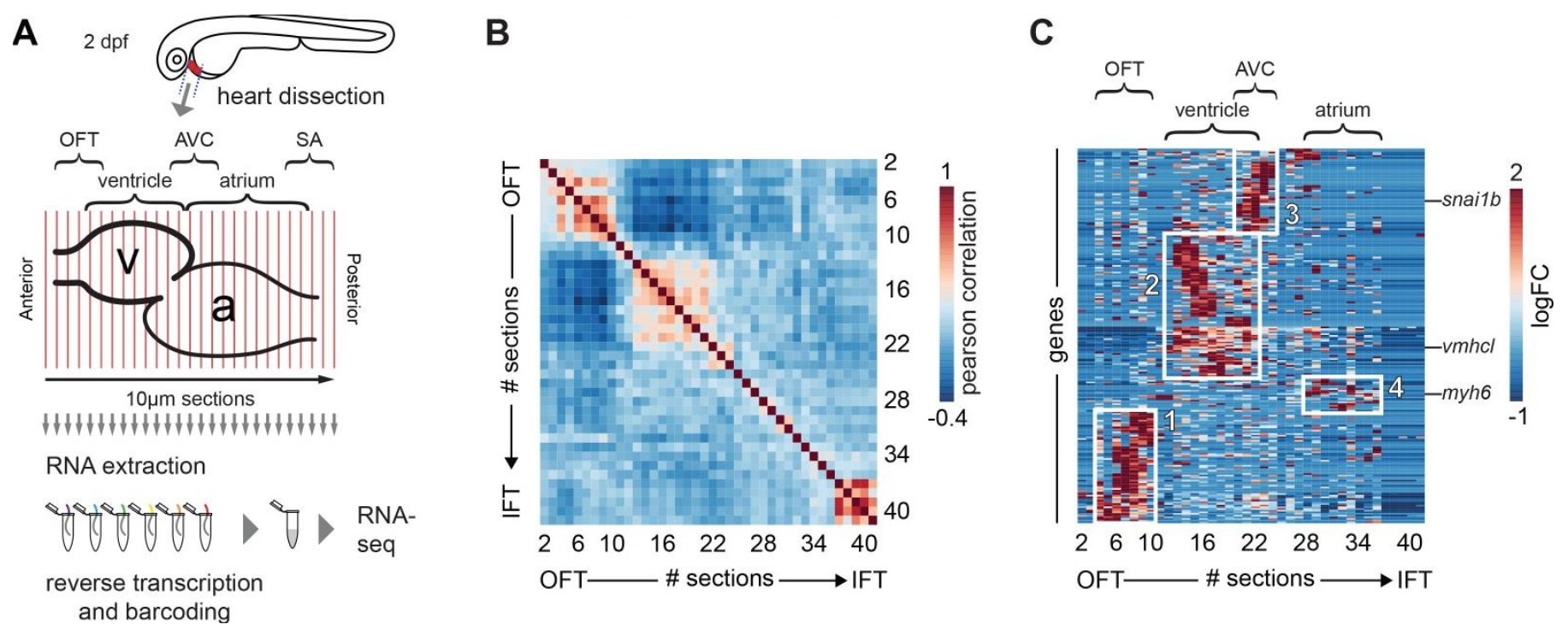
Development of specialized cell types and structures in the vertebrate heart is regulated by spatially-restricted molecular pathways. Disruptions in these pathways can cause severe congenital cardiac malformations or functional defects. To better understand these pathways and how they regulate cardiac development and function we used tomo-seq, combining high-throughput RNA sequencing with tissue sectioning, to establish a genome-wide expression dataset with high spatial resolution for the developing zebrafish heart. Analysis of the dataset revealed over 1100 genes differentially expressed in sub-compartments. Pacemaker cells in the sinoatrial region induce heart contractions, but little is known about the mechanisms underlying their development and function. Using our transcriptome map, we identified spatially restricted Wnt/β-catenin signaling activity in pacemaker cells, which was controlled by Islet-1 activity. Moreover, Wnt/β-catenin signaling at a specific developmental stage in the myocardium controls heart rate by regulating pacemaker cellular response to parasympathetic stimuli. Thus, this high-resolution transcriptome map incorporating all cell types in the embryonic heart can expose spatially-restricted molecular pathways critical for specific cardiac functions. |
| Key word |
Islet-1; RNA-seq; developmental biology; heart; pacemaker; stem cells; wnt; zebrafish |
| Publication |
Burkhard SB, Bakkers J. Spatially resolved RNA-sequencing of the embryonic heart identifies a role for Wnt/β-catenin signaling in autonomic control of heart rate. Elife 2018 Feb 5;7. PMID: 29400650 |
| Abstract |
Development of specialized cells and structures in the heart is regulated by spatially -restricted molecular pathways. Disruptions in these pathways can cause severe congenital cardiac malformations or functional defects. To better understand these pathways and how they regulate cardiac development we used tomo-seq, combining high-throughput RNA-sequencing with tissue-sectioning, to establish a genome-wide expression dataset with high spatial resolution for the developing zebrafish heart. Analysis of the dataset revealed over 1100 genes differentially expressed in sub-compartments. Pacemaker cells in the sinoatrial region induce heart contractions, but little is known about the mechanisms underlying their development. Using our transcriptome map, we identified spatially restricted Wnt/β-catenin signaling activity in pacemaker cells, which was controlled by Islet-1 activity. Moreover, Wnt/β-catenin signaling controls heart rate by regulating pacemaker cellular response to parasympathetic stimuli. Thus, this high-resolution transcriptome map incorporating all cell types in the embryonic heart can expose spatially restricted molecular pathways critical for specific cardiac functions. |