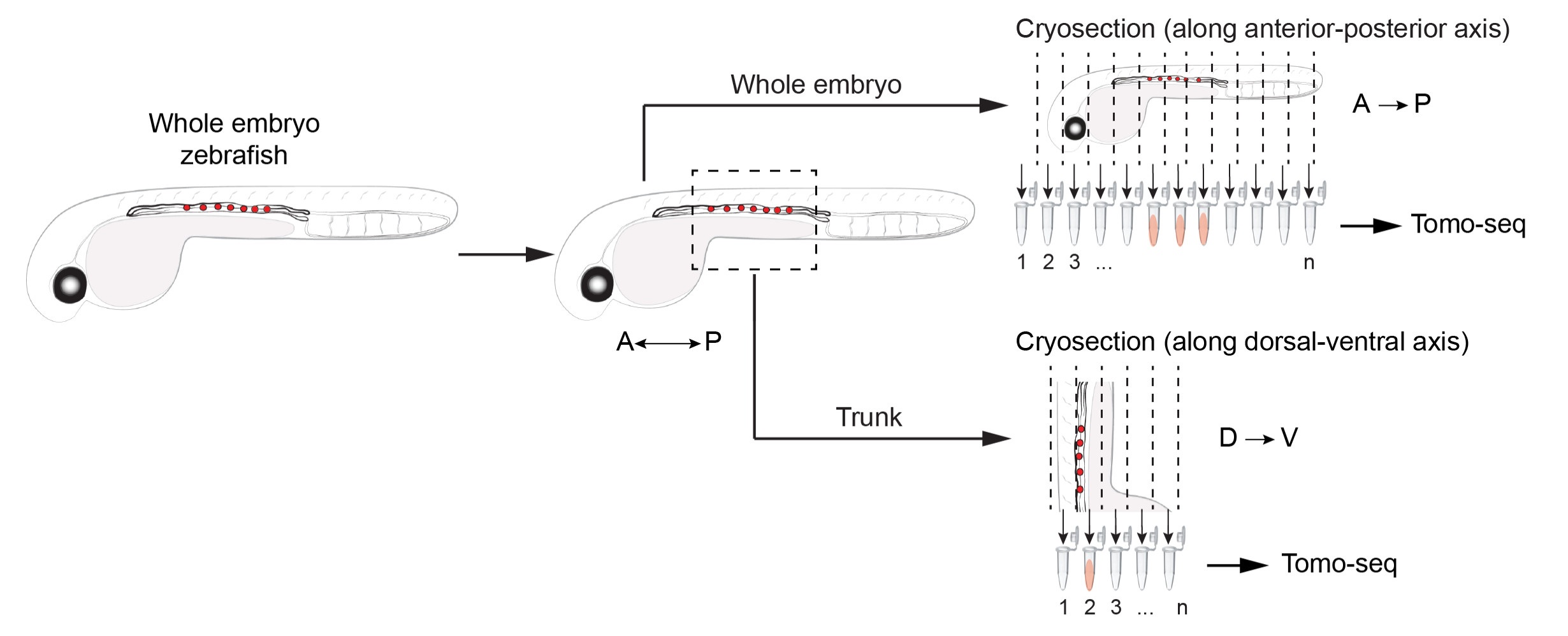
|
Dataset ID |
Tissue / Organ |
Technique |
Experiment type |
Sample description |
Source dataset ID |
| 1. |
GSE128350 (1.67dpf whole embryo replicate1) |
embryo |
Tomo-seq |
baseline |
late embryo, 1.67dpf, whole embryo, untreatment |
GEO: GSM3672145
|
| 2. |
GSE128350 (1.17dpf whole embryo replicate1) |
embryo |
Tomo-seq |
baseline |
late embryo, 1.17dpf, whole embryo, untreatment |
GEO: GSM3672146
|
| 3. |
GSE128350 (1.17dpf whole embryo replicate2) |
embryo |
Tomo-seq |
baseline |
late embryo, 1.17dpf, whole embryo, untreatment |
GEO: GSM3672147
|
| 4. |
GSE128350 (1.17dpf trunk) |
embryo |
Tomo-seq |
baseline |
late embryo, 1.17dpf, trunk, untreatment |
GEO: GSM3672148
|
| 5. |
GSE128350 (1.67dpf trunk) |
embryo |
Tomo-seq |
baseline |
late embryo, 1.67dpf, trunk, untreatment |
GEO: GSM3672149
|
| 6. |
GSE128350 (1.67dpf whole embryo replicate2) |
embryo |
Tomo-seq |
baseline |
late embryo, 1.67dpf, whole embryo, untreatment |
GEO: GSM3672157
|
| 7. |
GSE128350 (1.17dpf whole embryo replicate3) |
embryo |
Tomo-seq |
baseline |
late embryo, 1.17dpf, whole embryo, untreatment |
GEO: GSM3672158
|