OVERVIEW
FishSCT is a fish single-cell transcriptome database, with Danio rerio as the primary study subject. We collected 129 datasets from 44 projects from GEO and SRA common platforms and conducted unified analysis through a pipeline.
Finally, 964 / 26,965 marker / potential marker gene information of 245 cell type and a large number of expression profiles with single cell resolution were obtained. Among them, Danio rerio datasets are up to 117, covering 25 tissues at 36 time points in the growth and development stage of Danio rerio. In addition to Danio rerio, we also included the scRNA-seq data of 8 fish species to enable users to conduct comparative analysis between species.
Cite: Cheng Guo, Weidong Ye, Mijuan Shi et al. FishSCT: a zebrafish-centric database for exploration and visualization of fish single-cell transcriptome. Science China Life Science (2023). 10.1007/s11427-022-2293-4.
STATISTICS OF SPECIES
FishSCT stored 848 / 13,800 marker / potential marker information for 222 cell types of Danio rerio, and it's datasets cover 25 tissue types at 36 time points in the growth and development stage. The statistics of cell types and markers in each tissue are shown in the figure below left.
The figure on the below right is a drill-down pie chart, showing the marker proportion information of tissue and cell types of Danio rerio. When users click on the sector area, the figure will be switched to the next level to show the marker proportion of cell types contained in the tissue.
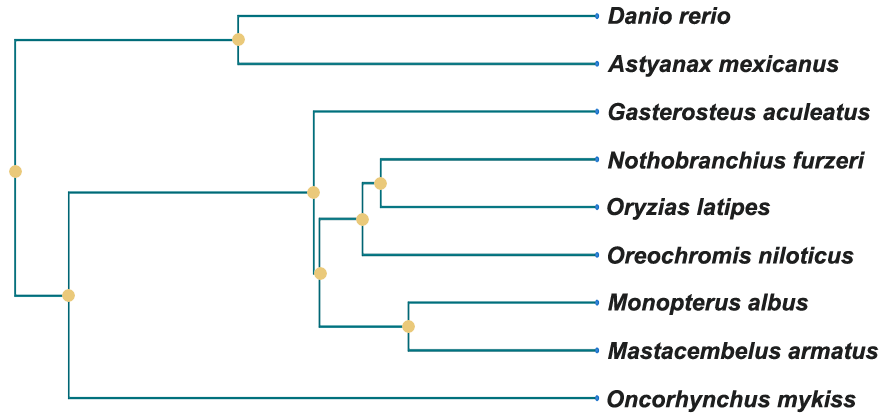
The phylogenetic relationships among the ray-finned fishes in FishSCT are shown in the figure below left. In addition to Danio rerio, Oryzias latipes is also the model species. FishSCT also includes scRNA-seq data for several economically important fish species.
FishSCT stored 116 / 13,165 marker / potential marker information for 59 cell types of other 8 fishes. The statistics of cell types and markers in each fish are shown in the following figure on the right.
BASIC INFORMATION OF SPECIES
model fish

Genome version:
>
GRCz11
Transcriptome source:
>
ensembl
Bioproject source:
>
GEO
>
SRA
zebrafish (Danio rerio)
Classification:
Actinopteri (ray-finned fishes) > Cypriniformes (Carps) > Danionidae (Danios) > Danioninae
ornamental fish

Bioproject source:
>
SRA
Experiment type: baseline
Source dataset id:
SRR11886701, SRR11917464, SRR15422711,
SRR15422691, SRR15422671, SRR15422704,
SRR15422680, SRR15422698, SRR15422699,
SRR15422703
mexican tetra (Astyanax mexicanus)
Classification:
Actinopteri (ray-finned fishes) > Characiformes (Characins) > Characidae (Characins; tetras) > Stethaprioninae
ecological fish

Transcriptome source:
>
ensembl
Bioproject source:
>
SRA
Experiment type:
> baseline
Source dataset id:
> SRR17665713
three-spined stickleback (Gasterosteus aculeatus)
Classification:
Actinopteri (ray-finned fishes) > Perciformes/Gasterosteoidei (Sticklebacks) > Gasterosteidae (Sticklebacks and tubesnouts)
ornamental fish

Bioproject source:
>
SRA
Experiment type:
> regeneration
Source dataset id:
> SRR11886701, SRR11917464
turquoise killifish (Nothobranchius furzeri)
Classification:
Actinopteri (ray-finned fishes) > Cyprinodontiformes (Rivulines, killifishes and live bearers) > Nothobranchiidae (African rivulines)
model fish

Transcriptome source:
>
ensembl
Bioproject source:
>
GEO
Experiment type:
> baseline
Source dataset id:
> GSM4959937, GSM4959938
Japanese medaka (Oryzias latipes)
Classification:
Actinopteri (ray-finned fishes) > Beloniformes (Needle fishes) > Adrianichthyidae (Ricefishes) > Oryziinae
economic fish

Bioproject source:
>
SRA
Experiment type:
> baseline
Source dataset id:
> SRR16095434
Nile tilapia (Oreochromis niloticus)
Classification:
Actinopteri (ray-finned fishes) > Cichliformes (Cichlids, convict blennies) > Cichlidae (Cichlids) > Pseudocrenilabrinae
economic fish

Transcriptome source:
>
ensembl
Bioproject source:
>
GEO
Experiment type:
> baseline
Source dataset id:
> GSM4661001
swamp eel (Monopterus albus)
Classification:
Actinopteri (ray-finned fishes) > Synbranchiformes (Spiny eels) > Synbranchidae (Swamp-eels)
economic fish

Bioproject source:
>
GEO
Experiment type:
> baseline
Source dataset id:
> GSM5333024
zig-zag eel (Mastacembelus armatus)
Classification:
Actinopteri (ray-finned fishes) > Synbranchiformes (Spiny eels) > Mastacembelidae (Spiny eels)
economic fish

Transcriptome source:
>
ensembl
Bioproject source:
>
GEO
Experiment type:
> baseline
Source dataset id:
> GSM4792223, GSM4792224, GSM4792225
rainbow trout (Oncorhynchus mykiss)
Classification:
Actinopteri (ray-finned fishes) > Salmoniformes (Salmons) > Salmonidae (Salmonids) > Salmoninae

Danio rerio
Genome version:
Transcriptome source:

Astyanax mexicanus
Genome version:
Transcriptome source:
Experiment type: baseline
Source dataset id:
SRR11886701, SRR11917464, SRR15422711, SRR15422691, SRR15422671, SRR15422704, SRR15422680, SRR15422698, SRR15422699, SRR15422703

Gasterosteus aculeatus
Genome version:
Transcriptome source:
Experiment type:
> baseline
Source dataset id:
> SRR17665713

Nothobranchius furzeri
Genome version:
Transcriptome source:
Experiment type:
> regeneration
Source dataset id:
> SRR11886701, SRR11917464

Oryzias latipes
Genome version:
Transcriptome source:
Experiment type:
> baseline
Source dataset id:
> GSM4959937, GSM4959938

Oreochromis niloticus
Genome version:
Transcriptome source:
Experiment type:
> baseline
Source dataset id:
> SRR16095434

Monopterus albus
Genome version:
Transcriptome source:
Experiment type:
> baseline
Source dataset id:
> GSM4661001

Mastacembelus armatus
Genome version:
Transcriptome source:
Experiment type:
> baseline
Source dataset id:
> GSM5333024

Oncorhynchus mykiss
Genome version:
Transcriptome source:
Experiment type:
> baseline
Source dataset id:
> GSM4792223, GSM4792224, GSM4792225